Evolution and Development
For the past 25 years the Bier lab has studied how secreted proteins known as morphogens including BMPs subdivide the dorsal-ventral axis of the fruit fly embryo into neural versus epidermal regions and how such processes result in the formation of sharp boundaries in the embryo and in primordia for adult structures such as wings. BMP mediated patterning in the embryo is among the most conserved of evolutionary processes.
|
BMP Patterning of the Embryonic DV axis
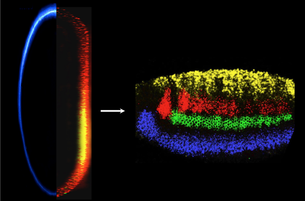
We first cloned the Drosophila short gastrulation gene (sog) (Francois et al., 1994), noted that it was orthologous to the vertebrate neural inducer chordin (Francois and Bier, 1995; Schmidt et al., 1995), and showed that it blocks an autoactivating feedback loop through which BMPs are poised to spread from the epidermal ectoderm into the neural ectoderm (Biehs et al., 1996). We demonstrated that Sog diffuses from the lateral neuroectoderm to form a ventral-to-dorsal concentration gradient within the epidermis (Srinivasan et al., 2002) that creates and inverse BMP activity gradient required to subdivide that region into two domains. The large extracellular Sog protein is also processed into several distinct BMP inhibitory forms (Yu et al. 2000, 2004; Araujo et al., 2003; Bier 2008).
|
|
We also collaborated with the Arthur Lander’s group (UCI) to mathematically model the Sog-BMP interaction in the dorsal ectoderm (Mizutani et al., 2005). Reciprocally, BMPs also diffuse ventrally into the neuroectoderm to partition the neuroectoderm into discrete regions of the future CNS (Mizutani et al. 2006; Mizutani and Bier, 2008). Since BMPs also play a role in CNS patterning in vertebrates, we collaborated with the Fisher (U. Penn) and Pyrowolakis (Univ. Freiburg) to ask whether this patterning is accomplished by conserved or distinct mechanisms in flies versus fish (Esteves et al., 2011). We found that BMPs produce the same output patterns in the CNS by opposite mechanisms (dose-dependent gene repression in flies versus graded gene activation in fish), highlighting the flexibility of evolutionary mechanisms despite stringent constraints on function (reviewed in Mizutani and Bier, 2008; Bier, 2011; and De Robertis, 2015).
|
|
Wing Vein Patterning
In our first studies we used gene markers to visualize developing vein primordia and showed that they form in response to localized EGF-R signaling (Sturtevant et al., 1993; Guichard et al., 1999), which then engages BMPs in a positive feedback loop (Yu et al., 1996). We have proposed that veins form at the borders of broad gene expression domains via an “for export-only” form of signaling in which one group of cells produces a short range signal to which they cannot respond themselves (Sturtevant and Bier, 1995; Sturtevant et al.,1997). We provided direct evidence for this hypothesis regarding formation of the L2 and L5 vein primordia which respectively express the knirps (kni) gene along the anterior border of the spalt expression domain (Lunde et al., 1998) and the abrupt (ab) gene posterior edge of the omb domain (Cook et al., 2004). We identified the cis regulatory module (CRM) directing L2 expression of the kni in the L2 primordium (Lunde et al., 2003) and are currently using CRISPR mutagenesis to analyze function of the L2-CRM in its native chromosomal context. We are also examining evolution of vein patterning by analyzing cis-regulation of this gene in flies (M. abdita) that have an extra vein relative to Drosophila that runs along the posterior border of the spalt domain. Genetic analysis of this vein patterning process in distantly related fly stimulated us to devise the MCR method that forms the basis for Active Genetics method which is described in a separate section. The wing is also a useful system for analyzing human disease gene function (Disease Mechanism section).
|
|
Technology Development
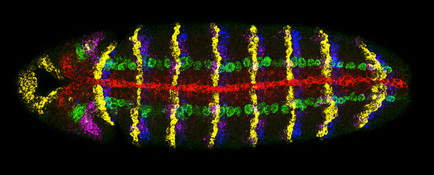
We have also contributed to development of new genetic and imaging technologies and bioinformatic tools (see below). As an extension of the PIs former efforts as one of the original developers of the enhancer trap method we devised enhancer piracy as a strategy to score directly for gene mis-expression phenotypes (Noll et al., 1994) and developed a highly efficient genetic method to screen for dominant negative of gain-of-function mutations in genes of interest (Guichard et al., 2002). We also developed methods for multi-label in situ hybridization (O’Neill and Bier, 1994) culminating a robust and now broadly used multiplex fluorescence method (developed in collaboration with the McGinnis lab) that permits up to seven different gene expression patterns to be visualized in a single embryo (Kosman et al., 2004) and collaborated with Terry Hwa’s group to generate quantitative imaging methods and data analysis tools that permit cell-by-cell analysis of gene expression levels during development (McHale et al., 2011).
|