Human Disease Mechanisms
The Bier lab uses the common fruit fly to study mechanisms of human disease. Our work in this area has focused recently on understanding the mechanisms by which bacterial toxins contribute to breaching host barriers. Thus, two toxins produced by anthrax bacteria trigger potentially fatal vascular leakage while cholera toxin leads to breakdown of the intestinal barrier leading to acute life-threatening diarrhea.
|
Closing the Loop
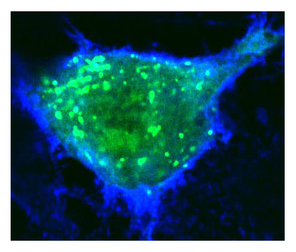
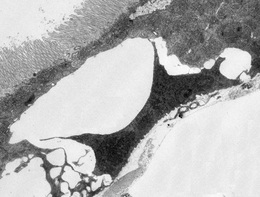
We carried out the first systematic analysis of human disease genes in Drosophila (Reiter et al., 2001 - see Bioinformatics section below), and have used flies to identify the Rho GEF Pebble as a candidate target of the ubiquitin E3 ligase disrupted in Angelman syndrome (Reiter et al., 2006), to discover a potent genetic interaction between over-expression of DSACM and COL6A2 as a potential contributing factor to the development of congenital heart defects in Down syndrome (Grossman et al., 2011), and to characterize Sestrins as negative regulators of TOR pathway signaling in metabolic homeostasis (Lee et al., 2010; 2012). Our current focus is to use flies to study host-pathogen interactions to gain mechanistic insights into the cell biological effects of bacterial toxins. For example, we have found that the anthrax toxins Lethal Factor (LF), a metalloprotease that cleaves and inactivated MEKKs and Edema Factor (EF), a highly active adenylate cyclase, cooperate to inhibit endocytic trafficking of proteins such as cadherins to cell-cell junctions in flies, mice,and human cells (Guichard et al., 2010, 2012). In mice, EF plays a key role in weakening vascular barrier integrity leading to severe vascular leakage, which is a common cause of death in late anthrax infection.
|
|
Similarly, we showed that cholera toxin, which like EF induces cAMP over-production, blocks junctional transport in the gut of flies, mice, and human cells, which contributes to the profuse life-threatening fluid loss associated with this disease (Guichard et al., 2013, 2014). We are currently analyzing the mechanisms by which other barrier disruptive toxins function using the Drosophila gut as an initial discovery model, and are conducting screens for compounds that can restore junctional trafficking and might be used to combat anthrax, cholera, as well as inflammatory diseases involving disruption of barrier integrity. Our collaborative studies to develop gene-drive systems to combat vector borne diseases described in the Active Genetics section synergize with these therapeutic efforts.
|
Bioinformatics
In parallel with our development of new genetic and imaging methods, we have contributed to developing a variety of new bioinformatics and quantitative analysis tools to aid in our experimental studies. With regard to our human disease project, we collaborated with Michael Gribskov (then at UCSD) to generate the still functional web-based tool Homophila, which serves as a simple but powerful portal for identifying fly homologs of human disease genes (Reiter et al., 2001). We then developed web-based tools to facilitate cross-genomic analysis of gene sequences (the Negative Proteome - Reiter et al., 2007) and a more general and powerful comparative tool for performing Boolean comparisons between diverse data sets (Booly - Do et al., 2010). As mentioned above, we have also collaborated with UCSD colleagues to develop quantitative image analysis tools and with Arthur Lander at UCI to formulate and test quantitative dynamic models of BMP-mediated patterning in early Drosophila embryos (Mizutani et al., 2005), which we are currently extending with additional collaborators (e.g., Scott Fraser, Sergey Nuzhdin, Paul Marjoram) at USC to generate new statistical and imaging methods for deducing gene regulatory networks.